Gplots Library
Using Bioconductor
The current release of Bioconductor is version3.12; it works with R version4.0.3. Users of older R andBioconductor must update their installation to take advantageof new features and to access packages that have been added toBioconductor since the last release.
The development version of Bioconductor is version3.13; it works with R version4.1.0. More recent ‘devel’versions of R (if available) will be supported during the nextBioconductor release cycle.
Install the latest release of R, then get the latest version ofBioconductor by starting R and entering the commands
The above plot is only for demonstration purposes, and it shows some of the many customization options available in the ggplot2 library. For more options, please refer to the ggplot2 documentation. If you have any questions, please feel free to leave a comment or reach out to me on Twitter. Pastebin.com is the number one paste tool since 2002. Pastebin is a website where you can store text online for a set period of time.
It may be possible to change the Bioconductor version of an existinginstallation; see the ‘Changing version’ section of the BiocManagervignette.
Details, including instructions toinstall additional packages and toupdate,find, andtroubleshoot are providedbelow. A devel version ofBioconductor is available. There are goodreasons for using BiocManager::install() formanaging Bioconductor resources.
Install R
- Download the most recent version of R. The R FAQs and the RInstallation and Administration Manual contain detailed instructionsfor installing R on various platforms (Linux, OS X, and Windows beingthe main ones).
Start the R program; on Windows and OS X, this will usually meandouble-clicking on the R application, on UNIX-like systems, type“R” at a shell prompt.
As a first step with R, start the R help browser by typing
help.start()in the R command window. For help on anyfunction, e.g. the “mean” function, type? mean.
[ Back to top ]
Install Bioconductor Packages
To install core packages, type the following in an R command window:
Install specific packages, e.g., “GenomicFeatures” and “AnnotationDbi”, with
The install() function (in the BiocManager package) has arguments that changeits default behavior; type ?install for further help.
For a more detailed explanation on using BiocManager and its advanced usage,such as version switching, please refer to theBiocManager vignette.
[ Back to top ]
Find Bioconductor Packages
Visit the software package listto discover available packages.
To search through available packages programmatically, use the following:
For example, using a “^org” search pattern will show all of the availableorganism annotation packages.
[ Back to top ]
Update Installed Bioconductor Packages
Bioconductor packages, especially those in the development branch, areupdated fairly regularly. To identify packages requiring update withinyour version of Bioconductor, start a new session of R and enter
Use the argument ask=FALSE to update old packages without beingprompted. Read the help page for ?install for additional details.
Upgrading installed Bioconductor packages
Some versions of R support more than one version of Bioconductor. Touse the latest version of Bioconductor for your version of R, enter
Remember that more recent versions of Bioconductor may be available if yourversion of R is out-of-date.
For more details on Bioconductor approaches to versioning, seethe advanced sectionin the vignette and version numbering in the developer reference section.
Recompiling installed Bioconductor packages
Rarely, underlying changes in the operating system require ALLinstalled packages to be recompiled for source (C or Fortran)compatibility. One way to address this might be to start a new Rsession and enter
As this will reinstall all currently installed packages, it likelyinvolves a significant amount of network bandwidth and compilationtime. All packages are implicitly updated, and the cumulative effectmight introduce wrinkles that disrupt your work flow. It also requiresthat you have the necessary compilers installed.
[ Back to top ]
Troubleshoot Package Installations
Use the commands
to flag packages that are either out-of-date or too new for yourversion of Bioconductor. The output suggests ways to solve identifiedproblems, and the help page ?valid lists arguments influencingthe behavior of the function.
Troubleshoot BiocManager
One likely reason for BiocManager not working on your system couldbe that your version of R is too old for BiocManager. In orderavoid this issue, please ensure that you have the latest version of Rinstalled in your system. BiocManager supports R versions from 3.5.0and above.
[ Back to top ]
Why use BiocManager::install()?
BiocManager::install() is the recommended way to install Bioconductorpackages. There are several reasons for preferring this to the‘standard’ way in which R pacakges are installed viainstall.packages().
Bioconductor has a repository and release schedule that differs from R(Bioconductor has a ‘devel’ branch to which new packages and updatesare introduced, and a stable ‘release’ branch emitted once every 6months to which bug fixes but not new features are introduced).
A consequence of the mismatch between R and Bioconductor releaseschedules is that the Bioconductor version identified byinstall.packages() is sometimes not the most recent ‘release’available. For instance, an R minor version may be introduced somemonths before the next Bioc release. After the Bioc release the usersof the R minor version will be pointed to an out-of-date version ofBioconductor.
A consequence of the distinct ‘devel’ branch is thatinstall.packages() sometimes points only to the ‘release’repository, whereas Bioconductor developers and users wantingleading-edge features wish to access the Bioconductor ‘devel’repository. For instance, the Bioconductor 3.0 release is availablefor R.3.1.x, so Bioconductor developers and leading-edge users need tobe able to install the devel version of Bioconductor packages into thesame version (though perhaps different instance or at least librarylocation) of R that supports version 2.14 of Bioconductor.
An indirect consequence of Bioconductor’s structured release is thatpackages generally have more extensive dependencies with one another,both explicitly via the usual package mechanisms and implicitlybecause the repository, release structure, and Bioconductor communityinteractions favor re-use of data representations and analysisconcepts across packages. There is thus a higher premium on knowingthat packages are from the same release, and that all packages arecurrent within the release.
The BiocManager package serves as the primary way to ensure thatthe appropriate Bioconductor installation is used with respectto the version of R in use regardless of the R and Bioconductorrelease cycles.
The install() function is provided by BiocManager. This is awrapper around install.packages, but with the repository chosenaccording to the version of Bioconductor in use, rather than to theversion relevant at the time of the release of R.
install() also nudges users to remain current within a release, bydefault checking for out-of-date packages and asking if the user wouldlike to update
The BiocManager package provides facilities for switching to the‘devel’ version of Bioconductor
(at some points in the R / Bioconductor release cycle use of ‘devel’requires use of a different version of R itself, in which case theattempt to install devel fails with an appropriate message).
The BiocManager package also provides valid() to test that theinstalled packages are not a hodgepodge from different Bioconductorreleases (the ‘too new’ packages have been installed from sourcerather than a repository; regular users would seldom have these).
For users who spend a lot of time in Bioconductor, the featuresoutlined above become increasingly important and install() is muchpreferred to install.packages().
[ Back to top ]
Pre-configured Bioconductor
Bioconductor is also available as a set ofAmazon Machine Images (AMIs) andDocker images.
Legacy and Older R Versions
It is always recommended to update to the most current version of R andBioconductor. If this is not possible and R < 3.5.0 , please use the followingfor installing Bioconductor packages
To install core packages, type the following in an R command window:
Install specific packages, e.g., “GenomicFeatures” and “AnnotationDbi”, with
[ Back to top ]
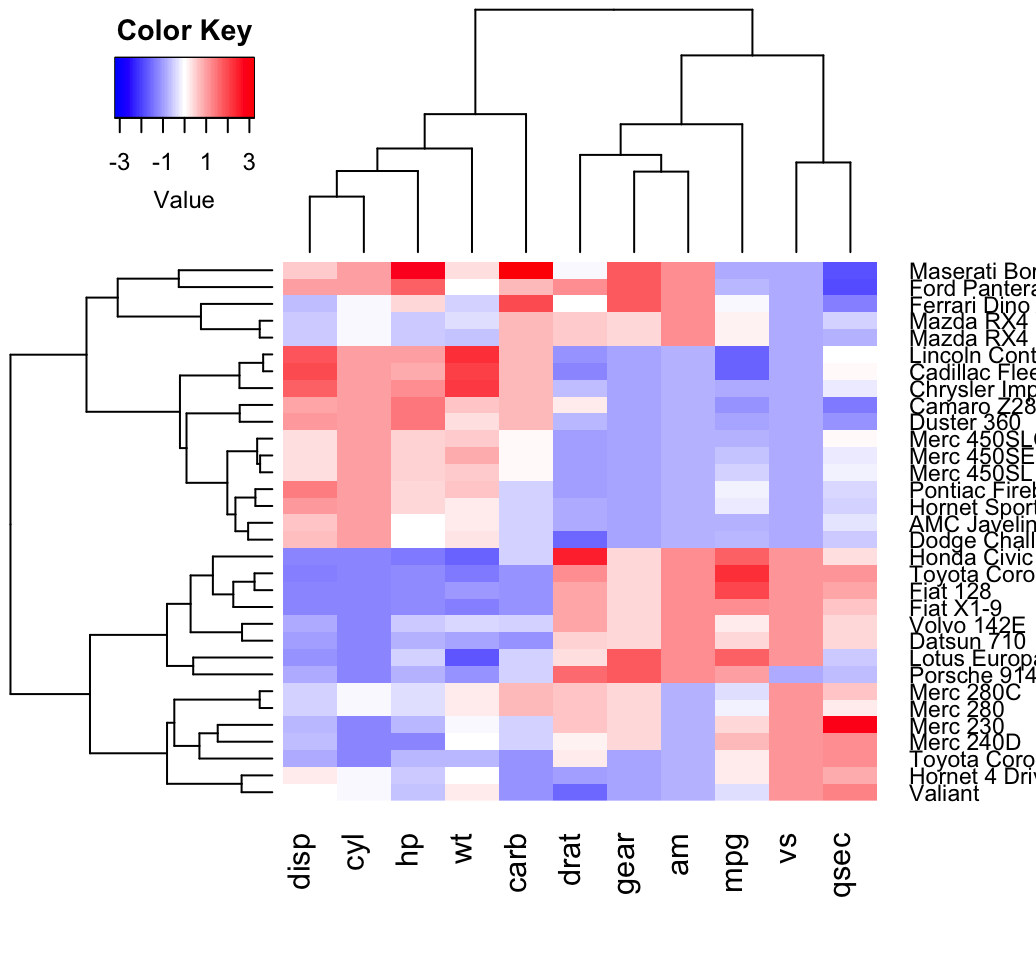
Two-Dimensional Visualization of Graphs
gplot produces a two-dimensional plot of graph g in collection dat. A variety of options are available to control vertex placement, display details, color, etc.
- Keywords
- graphs, hplot
Usage
Arguments
a graph or set thereof. This data may be valued.
integer indicating the index of the graph which is to be plotted. By default, g1.
String indicating the type of graph being evaluated. 'digraph' indicates that edges should be interpreted as directed; 'graph' indicates that edges are undirected; 'twomode' indicates that data should be interpreted as two-mode (i.e., rows and columns are distinct vertex sets). gmode is set to 'digraph' by default.
boolean indicating whether or not the diagonal should be treated as valid data. Set this true if and only if the data can contain loops. diag is FALSE by default.
a vector of vertex labels, if desired; defaults to the vertex index number.
user-specified vertex coordinates, in an NCOL(dat)x2 matrix. Where this is specified, it will override the mode setting.
boolean; should the output be jittered?
real number indicating the lower threshold for tie values. Only ties of value >thresh (by default in absolute value - see thresh.absval)are displayed. By default, thresh=0.
boolean; should the absolute value of edge weights be used when thresholding? (Defaults to TRUE; setting to FALSE leads to thresholding by signed weights.)
boolean; should arrows (rather than line segments) be used to indicate edges?
the vertex placement algorithm; this must correspond to a gplot.layout function.
boolean; should isolates be displayed?
boolean; should interactive adjustment of vertex placement be attempted?
boolean; if interactiveTRUE, should all vertices in the component be moved?
x axis label.
y axis label.
the x limits (min, max) of the plot.
the y limits of the plot.
amount to pad the plotting range; useful if labels are being clipped.
amount to pad label boxes (if boxed.labelsTRUE), in character size units.
boolean; should vertex labels be displayed?
boolean; place vertex labels within boxes?
position at which labels should be placed, relative to vertices. 0 results in labels which are placed away from the center of the plotting region; 1, 2, 3, and 4 result in labels being placed below, to the left of, above, and to the right of vertices (respectively); and label.pos>=5 results in labels which are plotted with no offset (i.e., at the vertex positions).
background color for label boxes (if boxed.labelsTRUE); may be a vector, if boxes are to be of different colors.
boolean; should vertices be enclosed within circles? (Can increase legibility for polygonal vertices.)
number of polygon sides for vertices; may be given as a vector, if vertices are to be of different types. By default, 50 sides are used (or 50 and 4, for two-mode data).
angle of rotation for vertices (in degrees); may be given as a vector, if vertices are to be rotated differently.
expansion factor for edge arrowheads.
character expansion factor for label text.
expansion factor for loops; may be given as a vector, if loops are to be of different sizes.

expansion factor for vertices; may be given as a vector, if vertices are to be of different sizes.
color for edges; may be given as a vector or adjacency matrix, if edges are to be of different colors.
color for vertex labels; may be given as a vector, if labels are to be of different colors.
color for vertices; may be given as a vector, if vertices are to be of different colors. By default, red is used (or red and blue, for two-mode data).
label border colors (if boxed.labelsTRUE); may be given as a vector, if label boxes are to have different colors.
border color for vertices; may be given as a vector, if vertex borders are to be of different colors.
line type for (positive weight) edges; may be given as a vector or adjacency matrix, if edges are to have different line types.
line type for negative weight edges, if any; may be given as per edge.lty.
line type for label boxes (if boxed.labelsTRUE); may be given as a vector, if label boxes are to have different line types.
line type for vertex borders; may be given as a vector or adjacency matrix, if vertex borders are to have different line types.
line width scale for edges; if set greater than 0, edge widths are scaled by edge.lwd*dat. May be given as a vector or adjacency matrix, if edges are to have different line widths.
line width for label boxes (if boxed.labelsTRUE); may be given as a vector, if label boxes are to have different line widths.
if uselenTRUE, curved edge lengths are scaled by edge.len.
if usecurveTRUE, the extent of edge curvature is controlled by edge.curv. May be given as a fixed value, vector, or adjacency matrix, if edges are to have different levels of curvature.
for curved edges (excluding loops), the number of line segments to use for the curve approximation.
for loops, the number of line segments to use for the curve approximation.
base length for plotting objects, as a fraction of the linear scale of the plotting region. Defaults to 0.01.
boolean; should we use edge.len to rescale edge lengths?
boolean; should we use edge.curve?
3d Scatter Plots Library For Processing
boolean; suppress plotting of axes?
boolean; plot vertices after plotting edges?
boolean; create a new plot? If newFALSE, vertices and edges will be added to the existing plot.
parameters to the gplot.layout function specified in mode.
additional arguments to plot.
Details
gplot is the standard network visualization tool within the sna library. By means of clever selection of display parameters, a fair amount of display flexibility can be obtained. Graph layout -- if not specified directly using coord -- is determined via one of the various available algorithms. These should be specified via the mode argument; see gplot.layout for a full list. User-supplied layout functions are also possible -- see the aforementioned man page for details.
Install Gplots Library In R
Note that where gmode'twomode', the supplied two-mode network is converted to bipartite form prior to computing coordinates (if not in that form already). vertex.col or other settings may be used to differentiate row and column vertices -- by default, row vertices are drawn as red circles, and column vertices are rendered as blue squares. If interactiveTRUE, then the user may modify the initial graph layout by selecting an individual vertex and then clicking on the location to which this vertex is to be moved; this process may be repeated until the layout is satisfactory. If interact.bycompTRUE as well, the vertex and all other vertices in the same component as that vertex are moved together.
Value
A two-column matrix containing the vertex positions as x,y coordinates.
References
Wasserman, S. and Faust, K. (1994) Social Network Analysis: Methods and Applications. Cambridge: Cambridge University Press.
See Also
plot, gplot.layout
Aliases
- gplot